Contents
clear all
close all
clc
s = RandStream('mt19937ar','Seed',sum(round(clock)));
RandStream.setGlobalStream(s);
1 Classic Efron - does Aspirin prevent heart attacks
nA = 11037;
nHA = 104;
nP = 11034;
nHP = 189;
nSamples = 1e5;
ratio = zeros(nSamples,1);
A1 = ones(nHA,1);
A2 = zeros(nA-nHA,1);
P1 = ones(nHP,1);
P2 = zeros(nP-nHP,1);
A = [A1;A2];
P = [P1;P2];
for ii = 1:nSamples
indicesA = randi(nA,[nA 1]);
indicesP = randi(nP,[nP 1]);
ratio(ii) = sum(A(indicesA))/sum(P(indicesP));
end
sortedRatio = sort(ratio);
lowerBoundIndex = round(nSamples/100*2.5);
upperBoundIndex = round(nSamples/100*97.5);
lowerBound = sortedRatio(lowerBoundIndex);
upperBound = sortedRatio(upperBoundIndex);
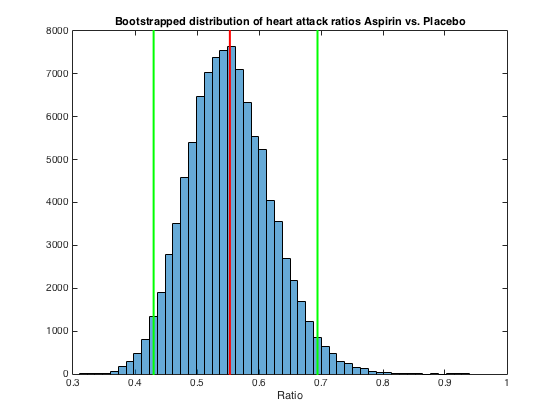
figure
histogram(ratio,50);
xlabel('Ratio')
line([mean(ratio) mean(ratio)], [min(ylim) max(ylim)],'color','r','linewidth',2)
line([lowerBound lowerBound], [min(ylim) max(ylim)],'color','g','linewidth',2)
line([upperBound upperBound], [min(ylim) max(ylim)],'color','g','linewidth',2)
title('Bootstrapped distribution of heart attack ratios Aspirin vs. Placebo')
2 Using resampling methods for cross-validation
numPoints = 8;
noiseMagnitude = 2;
x = -5:0.01:5;
y = x.^2 + noiseMagnitude .* randn(1,length(x));
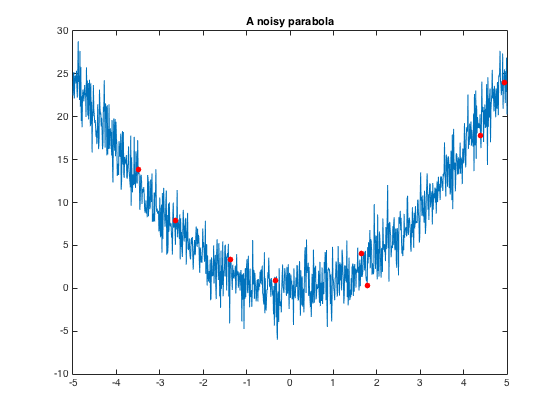
figure
plot(x,y)
title('A noisy parabola')
samplingIndices = randi(length(x),[numPoints,1 ]) ;
hold on;
plot(x(samplingIndices),y(samplingIndices),'.','color','r','markersize',20)
RMSE = [];
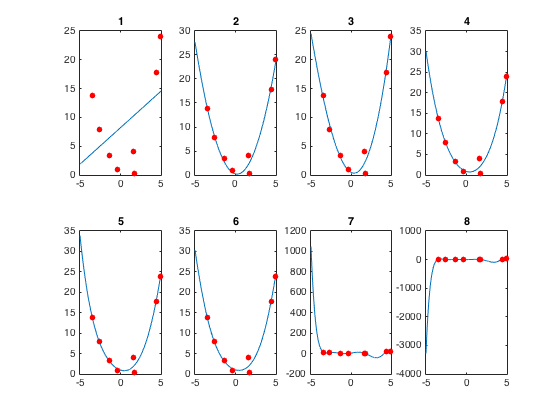
figure
for ii = 1:numPoints
subplot(2,4,ii)
p = polyfit(x(samplingIndices),y(samplingIndices),ii);
yHat = polyval(p,x);
plot(x,yHat)
hold on
plot(x(samplingIndices),y(samplingIndices),'.','color','r','markersize',20)
title(num2str(ii))
RMSE(ii) = sqrt(mean((y(samplingIndices)-yHat(samplingIndices)).^2));
end
figure
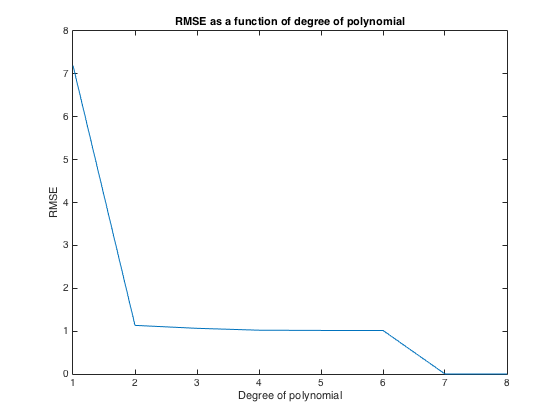
plot(1:numPoints,RMSE)
title('RMSE as a function of degree of polynomial')
xlabel('Degree of polynomial')
ylabel('RMSE')
numSamples = 1e2;
RMSE = zeros(numSamples,numPoints-1);
for ii = 1:numSamples
testIndex = randi(numPoints,1);
testSet = samplingIndices(testIndex);
trainingSet = samplingIndices;
trainingSet(testIndex,:) = [];
for jj = 1:numPoints-1
p = polyfit(x(trainingSet),y(trainingSet),jj);
yHat = polyval(p,x);
RMSE(ii,jj) = sqrt((y(testSet)-yHat(testSet)).^2);
end
end
figure
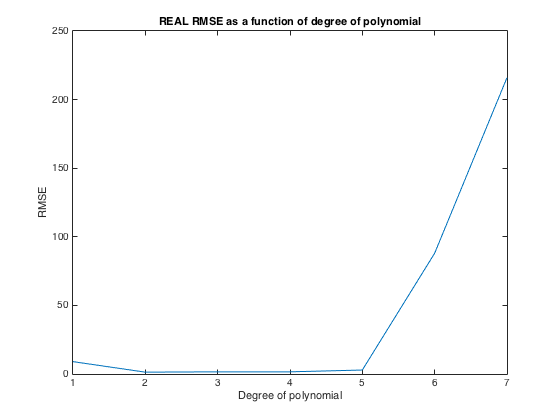
plot(1:numPoints-1,mean(RMSE))
title('REAL RMSE as a function of degree of polynomial')
xlabel('Degree of polynomial')
ylabel('RMSE')
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
Warning: Polynomial is not unique; degree >= number of data points.
3 Shortcut - stability of sample mean by hand and with MATLAB's built in bootstrp function
nRepeats = 1e3;
y = random('unif',0,1,[100 1])
m1 = zeros(nRepeats,1);
for ii = 1:nRepeats
indices = randi(length(y),[length(y) 1]) ;
m1(ii,1) = mean(y(indices));
end
m2 = bootstrp(nRepeats, @mean, y);
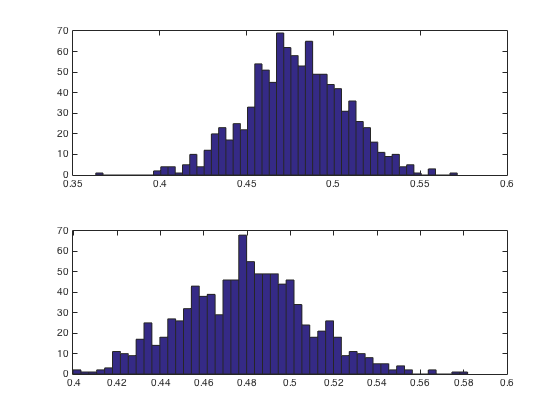
figure
subplot(2,1,1)
hist(m1,50)
subplot(2,1,2)
hist(m2,50)
mean(m1)
mean(m2)
std(m1)
std(m2)
y =
0.0076
0.1253
0.7569
0.2094
0.4729
0.0316
0.9912
0.2786
0.8140
0.4911
0.3587
0.4710
0.4195
0.5091
0.6741
0.7100
0.4897
0.5014
0.0007
0.3830
0.3474
0.6258
0.9185
0.9014
0.6600
0.8927
0.0038
0.4455
0.5090
0.3830
0.7089
0.6247
0.8909
0.2380
0.0854
0.1631
0.0758
0.4118
0.4839
0.6714
0.1564
0.6374
0.5745
0.5900
0.2622
0.9397
0.6651
0.8160
0.7631
0.8537
0.6834
0.9245
0.8710
0.1642
0.9891
0.1098
0.1353
0.9855
0.0413
0.5222
0.1700
0.9170
0.2860
0.6094
0.3714
0.0693
0.5561
0.0323
0.4145
0.8976
0.5232
0.2807
0.2814
0.5201
0.3245
0.4115
0.6242
0.6617
0.3796
0.6700
0.8352
0.0667
0.4345
0.1021
0.0417
0.3144
0.2083
0.9490
0.7538
0.7655
0.1035
0.5466
0.5532
0.0749
0.8732
0.1990
0.2426
0.6686
0.6568
0.1033
ans =
0.4784
ans =
0.4791
ans =
0.0282
ans =
0.0288
4 Comparing means with permutation tests
K = [117 123 111 101 121]
C = [98 104 106 92 88]
numMCS = 1e5;
nK = length(K);
nC = length(C);
testStatistic = sum(K)-sum(C)
dTS = zeros(numMCS,1);
allData = [K C];
for ii=1:numMCS
indices = randperm(nK+nC);
tempDATA = allData(indices);
group1 = tempDATA(1:nK);
tempDATA(:,1:nK) = [];
group2 = tempDATA;
dTS(ii) = sum(group1)-sum(group2);
end
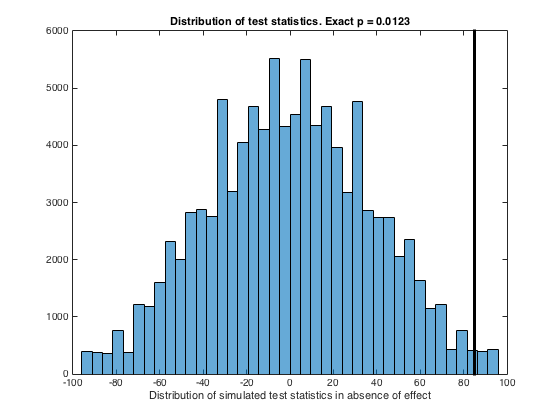
figure
histogram(dTS,40)
exactP = sum((dTS>=testStatistic))/length(dTS);
line([testStatistic testStatistic],[min(ylim) max(ylim)],'color','k','linewidth',3)
xlabel('Distribution of simulated test statistics in absence of effect')
title(['Distribution of test statistics. Exact p = ', num2str(exactP,'%1.4f')])
K =
117 123 111 101 121
C =
98 104 106 92 88
testStatistic =
85