Contents
- Now, let's actually use the formula we derived
- Let's explicitly add the distance between the two
- Now that we convinced ourselves that this is in fact the correct beta (geometrically)
- Multiple regression - simplest case: Adding a constant to the regression equation
- Let's plot the new regression line on top of the old figure
- Dropping marbles - which beta is the right beta?
- Going deeper into optimization - and gradient descent
- Gradient descent revisited
- PCA
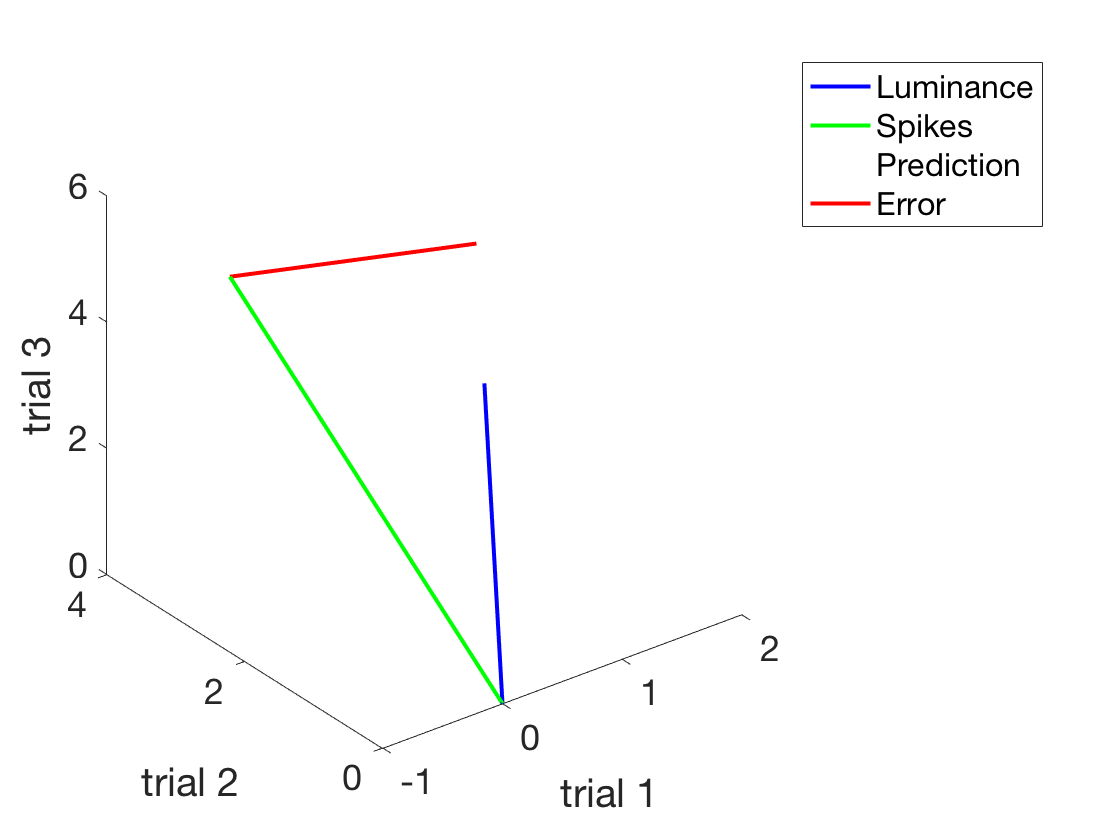
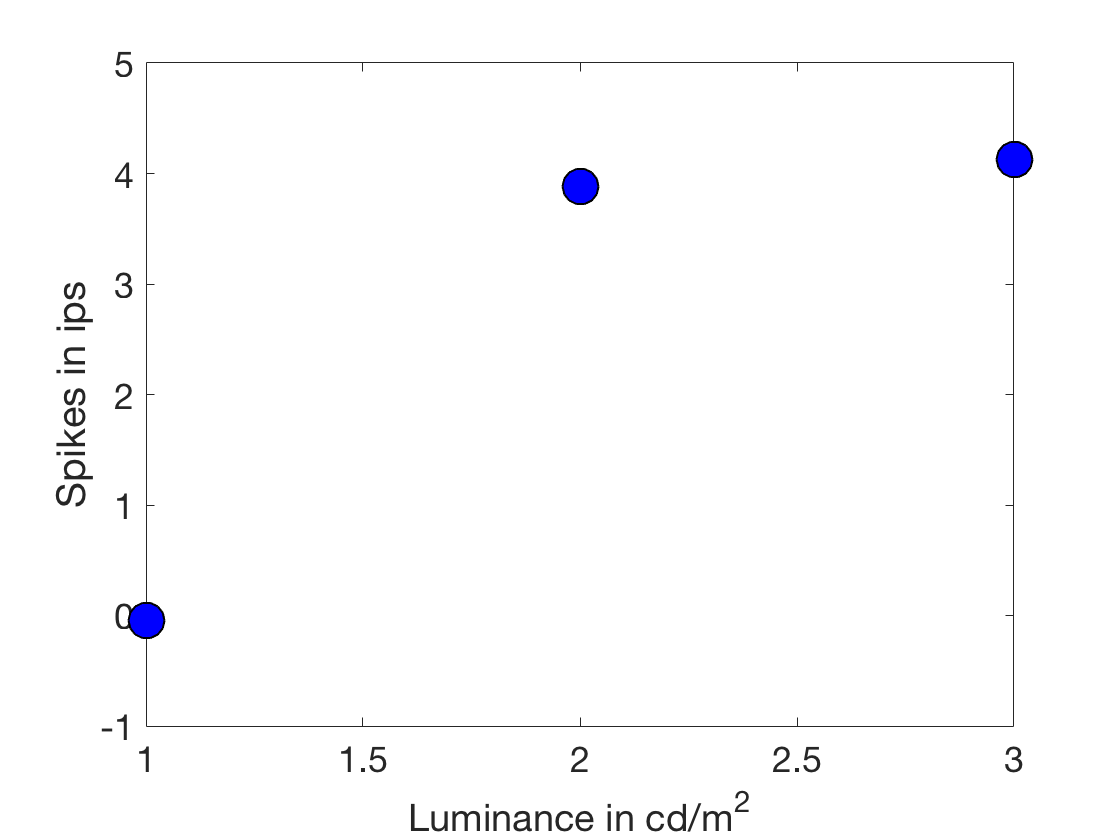
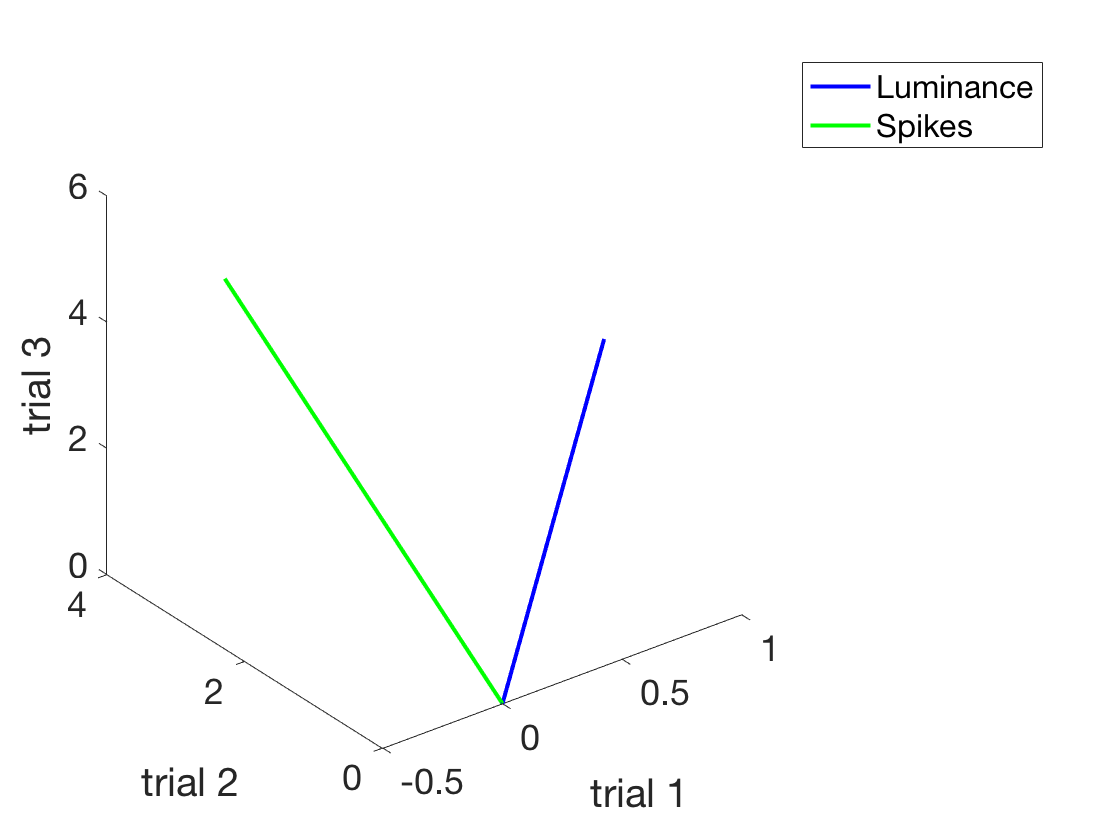
clear all close all xLuminance = [1:3]'; %A column vector of IV in cd/m^2 ySpikes = 1.5*xLuminance + 0.5 + randn(length(xLuminance),1).*2; %Noisy integrate and fire %Make a nice plot of the situation figure h = plot(xLuminance,ySpikes,'o') %First use of an object handle! set(h,'Markersize',18) h.MarkerFaceColor = 'b'; h.MarkerEdgeColor = 'k'; xlabel('Luminance in cd/m^2') ylabel('Spikes in ips') set(gca,'fontsize',18) %This is legit, but to see how the equation (which works) works %let's look at this graphically. Or geometrically. %To do that, we need to express the experiment visually. %Now, each trial is a dimension. figure h1 = plot3([0, xLuminance(1)], [0, xLuminance(2)], [0, xLuminance(3)]) h1.Color = 'b'; h1.LineWidth = 2; hold on h2 = plot3([0, ySpikes(1)], [0, ySpikes(2)], [0, ySpikes(3)]) h2.Color = 'g'; h2.LineWidth = 2; legend('Luminance','Spikes') xlabel('trial 1') ylabel('trial 2') zlabel('trial 3') set(gca,'fontSize',18) rotate3d on
h =
Line with properties:
Color: [0 0.447 0.741]
LineStyle: 'none'
LineWidth: 0.5
Marker: 'o'
MarkerSize: 6
MarkerFaceColor: 'none'
XData: [1 2 3]
YData: [-0.0428433530388914 3.88022557987611 4.13042332294946]
ZData: [1x0 double]
Use GET to show all properties
h1 =
Line with properties:
Color: [0 0.447 0.741]
LineStyle: '-'
LineWidth: 0.5
Marker: 'none'
MarkerSize: 6
MarkerFaceColor: 'none'
XData: [0 1]
YData: [0 2]
ZData: [0 3]
Use GET to show all properties
h2 =
Line with properties:
Color: [0.85 0.325 0.098]
LineStyle: '-'
LineWidth: 0.5
Marker: 'none'
MarkerSize: 6
MarkerFaceColor: 'none'
XData: [0 -0.0428433530388914]
YData: [0 3.88022557987611]
ZData: [0 4.13042332294946]
Use GET to show all properties
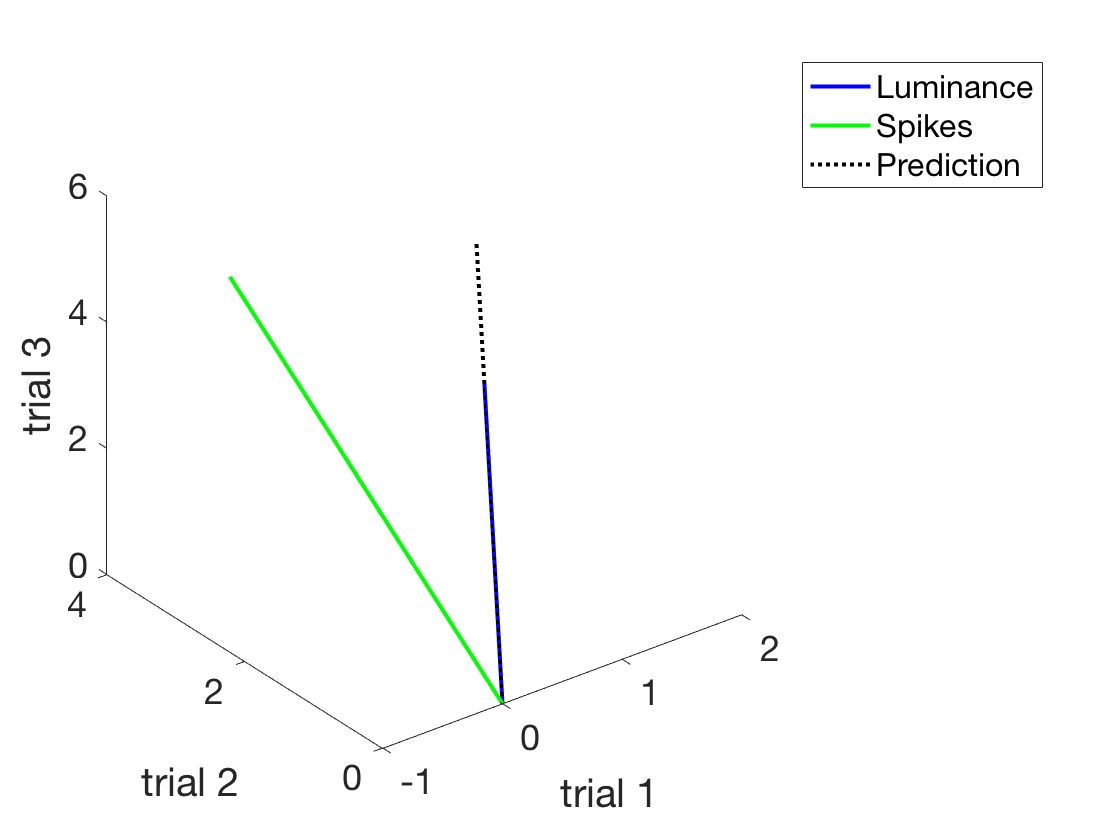
Now, let's actually use the formula we derived
beta = (ySpikes' * xLuminance)/(xLuminance'*xLuminance) %Find the beta beta = dot(ySpikes,xLuminance)/dot(xLuminance,xLuminance) %Same if you don't remember which is which prediction = beta*xLuminance %Make a prediction (simplest possible) %Add this to the plot - the plot thickens h3 = plot3([0, prediction(1)], [0, prediction(2)], [0, prediction(3)]); set(h3,'linewidth',2) set(h3,'linestyle',':') h3.Color = 'k' legend('Luminance','Spikes','Prediction') shg
beta =
1.43634841254012
beta =
1.43634841254012
prediction =
1.43634841254012
2.87269682508024
4.30904523762036
h3 =
Line with properties:
Color: [0 0 0]
LineStyle: ':'
LineWidth: 2
Marker: 'none'
MarkerSize: 6
MarkerFaceColor: 'none'
XData: [0 1.43634841254012]
YData: [0 2.87269682508024]
ZData: [0 4.30904523762036]
Use GET to show all properties
Let's explicitly add the distance between the two
h4 = plot3([ySpikes(1), prediction(1)], [ySpikes(2), prediction(2)], [ySpikes(3), prediction(3)]); set(h4,'linewidth',2) set(h4,'linestyle','-') h4.Color = 'r' legend('Luminance','Spikes','Prediction','Error') shg
h4 =
Line with properties:
Color: [1 0 0]
LineStyle: '-'
LineWidth: 2
Marker: 'none'
MarkerSize: 6
MarkerFaceColor: 'none'
XData: [-0.0428433530388914 1.43634841254012]
YData: [3.88022557987611 2.87269682508024]
ZData: [4.13042332294946 4.30904523762036]
Use GET to show all properties
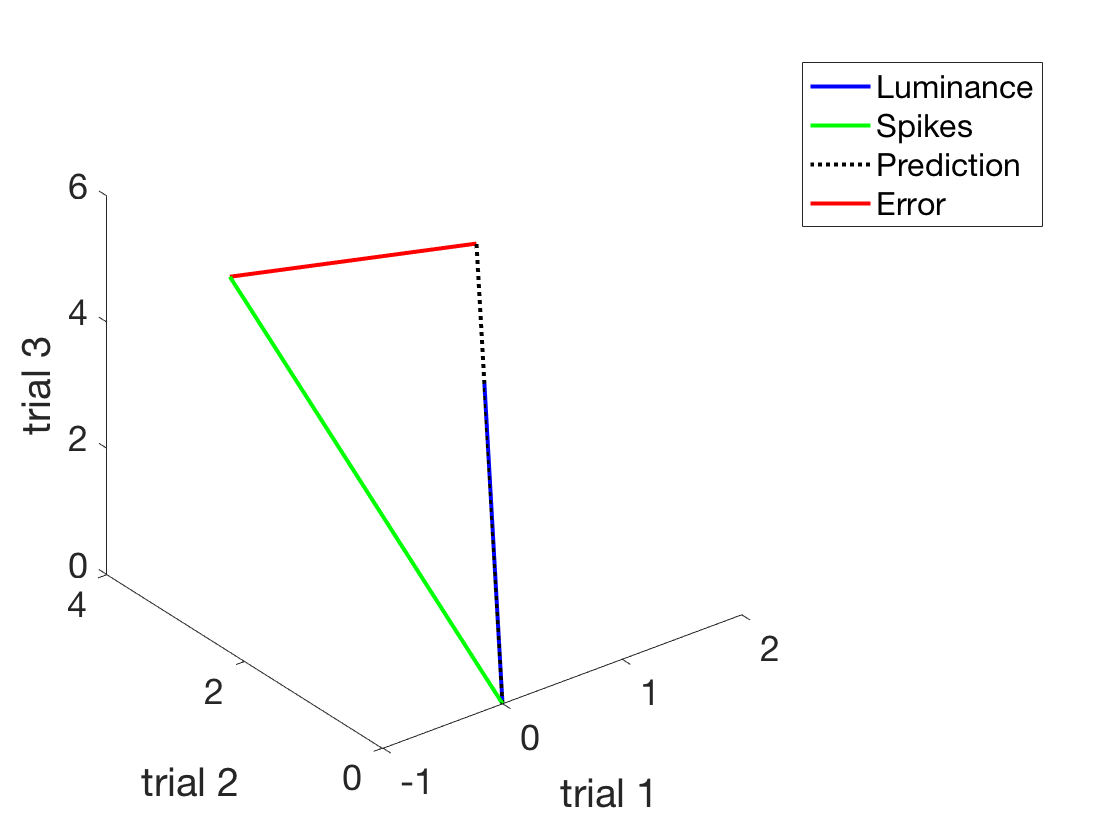
Now that we convinced ourselves that this is in fact the correct beta (geometrically)
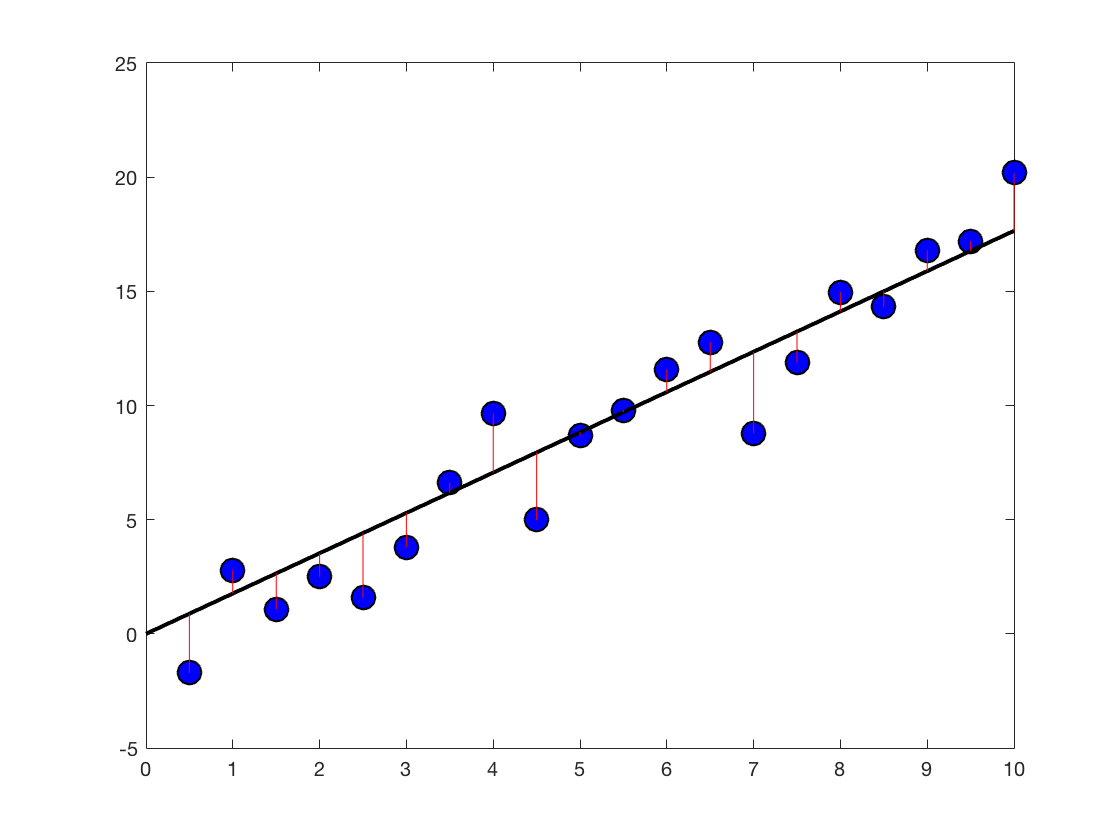
we can go back and plot the solution We could open the old figure again, but let's start from scratch What if we had 20 measurements (20 trials)?
maxLuminance = 10; xLuminance = [0.5:0.5:maxLuminance]'; %A column vector of IV in cd/m^2 - 20 luminance values ySpikes = 1.5*xLuminance + 0.5 + randn(length(xLuminance),1).*2; %Noisy integrate and fire beta = dot(ySpikes,xLuminance)/dot(xLuminance,xLuminance) %Same if you don't remember which is which prediction = beta*xLuminance %Make a prediction (simplest possible) figure h4 = plot(xLuminance,ySpikes,'o') set(h4,'Markersize',12) h4.MarkerFaceColor = 'b'; h4.MarkerEdgeColor = 'k'; pause hold on regressionLineX = linspace(0,maxLuminance,10); %Gives us 10 equally spaced numbers between 0 and 4. Intrapolation, x-base regressionLineY = beta .* regressionLineX; %Find the ys of the regression line h5 = plot(regressionLineX,regressionLineY,'k') set(h5,'linewidth',2) pause %h6 = plot(xLuminance,prediction,'o') %set(h6,'Markersize',18) %h6.MarkerFaceColor = 'k'; %h6.MarkerEdgeColor = 'y'; lh = line([xLuminance'; xLuminance'],[prediction'; ySpikes'], 'color','r'); pause
beta =
1.76497701782015
prediction =
0.882488508910077
1.76497701782015
2.64746552673023
3.52995403564031
4.41244254455039
5.29493105346046
6.17741956237054
7.05990807128062
7.9423965801907
8.82488508910077
9.70737359801085
10.5898621069209
11.472350615831
12.3548391247411
13.2373276336512
14.1198161425612
15.0023046514713
15.8847931603814
16.7672816692915
17.6497701782015
h4 =
Line with properties:
Color: [0 0.447 0.741]
LineStyle: 'none'
LineWidth: 0.5
Marker: 'o'
MarkerSize: 6
MarkerFaceColor: 'none'
XData: [1x20 double]
YData: [1x20 double]
ZData: [1x0 double]
Use GET to show all properties
h5 =
Line with properties:
Color: [0 0 0]
LineStyle: '-'
LineWidth: 0.5
Marker: 'none'
MarkerSize: 6
MarkerFaceColor: 'none'
XData: [1x10 double]
YData: [1x10 double]
ZData: [1x0 double]
Use GET to show all properties
Multiple regression - simplest case: Adding a constant to the regression equation
%Even having more than one predictor makes it a multiple regression, even %if it is a constant. %We need to represent the baseline %We need as many baselines as there are trials, and it always should have %the same value. Because we assume this is a constant baseline = ones(length(xLuminance),1) designMatrix = [xLuminance, baseline]; betas = pinv(designMatrix' * designMatrix) * designMatrix'*ySpikes; %We should get a vector of 2 betas out of this prediction = designMatrix*betas;
baseline =
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
Let's plot the new regression line on top of the old figure
regressionLineYBar = betas(1)*regressionLineX + betas(2) ... * ones(1,length(regressionLineX)); plot(regressionLineX,regressionLineYBar,'color','m','linewidth',2) pause
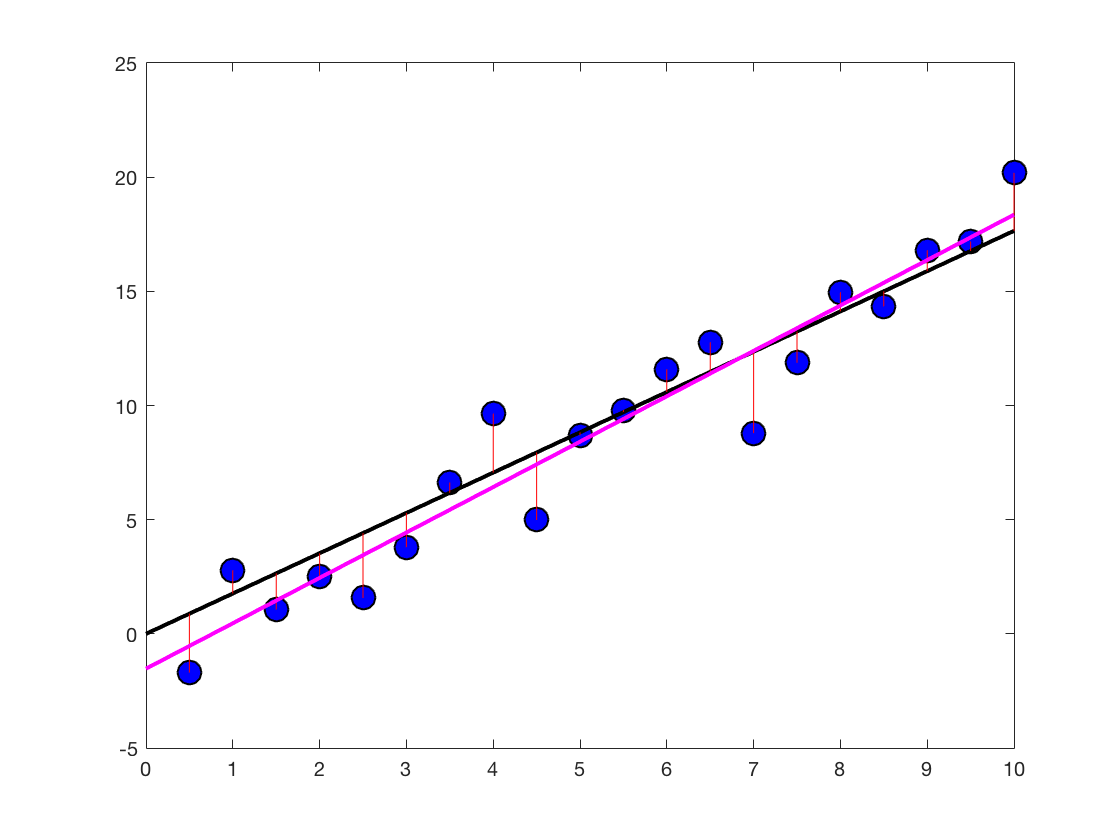
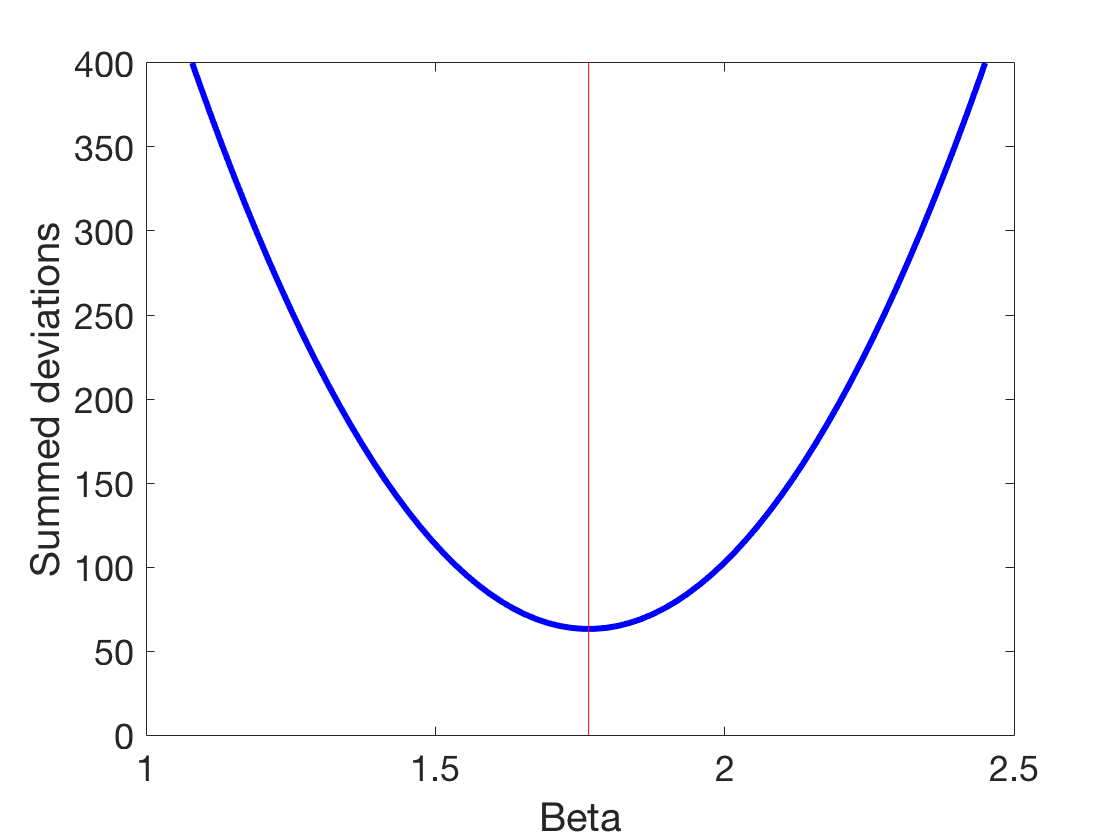
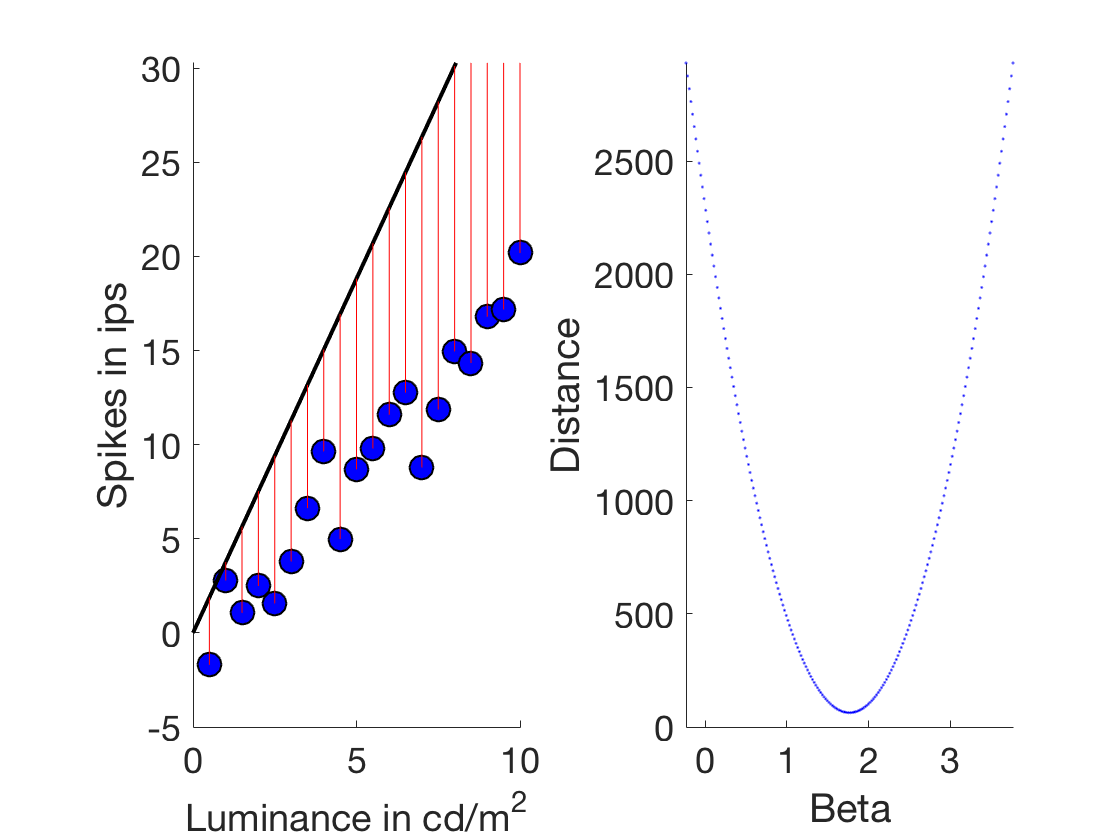
Dropping marbles - which beta is the right beta?
beta %Let's explore the space of betas startExploration = beta-2; endExploration = beta+2; numBeta = 200; testBetas = linspace(startExploration,endExploration,numBeta) ;%200 betas between the actual beta and +/- 2 %Let's just go through them and try them for ii = 1:numBeta prediction = testBetas(ii).*xLuminance; %Do this numBeta times %We now need to introduce a distance metric %We start with sum of squares (the most common maybe) distanceSum(ii) = sum((prediction-ySpikes).^2); %Sum of squares % distanceSum(ii) = sum((prediction-ySpikes)); %Simple summed deviations %distanceSum(ii) = sum(log(prediction-ySpikes)); %Absolute summed deviations end %Let's plot that figure plot(testBetas,distanceSum,'b','linewidth',3) %We also want to indicate with a line where the original beta was line([beta beta], [min(ylim) max(ylim)], 'color','r') xlabel('Beta') ylabel('Summed deviations') set(gca,'fontsize',18) set(gcf,'color','w') ylim([0 400])
beta =
1.76497701782015
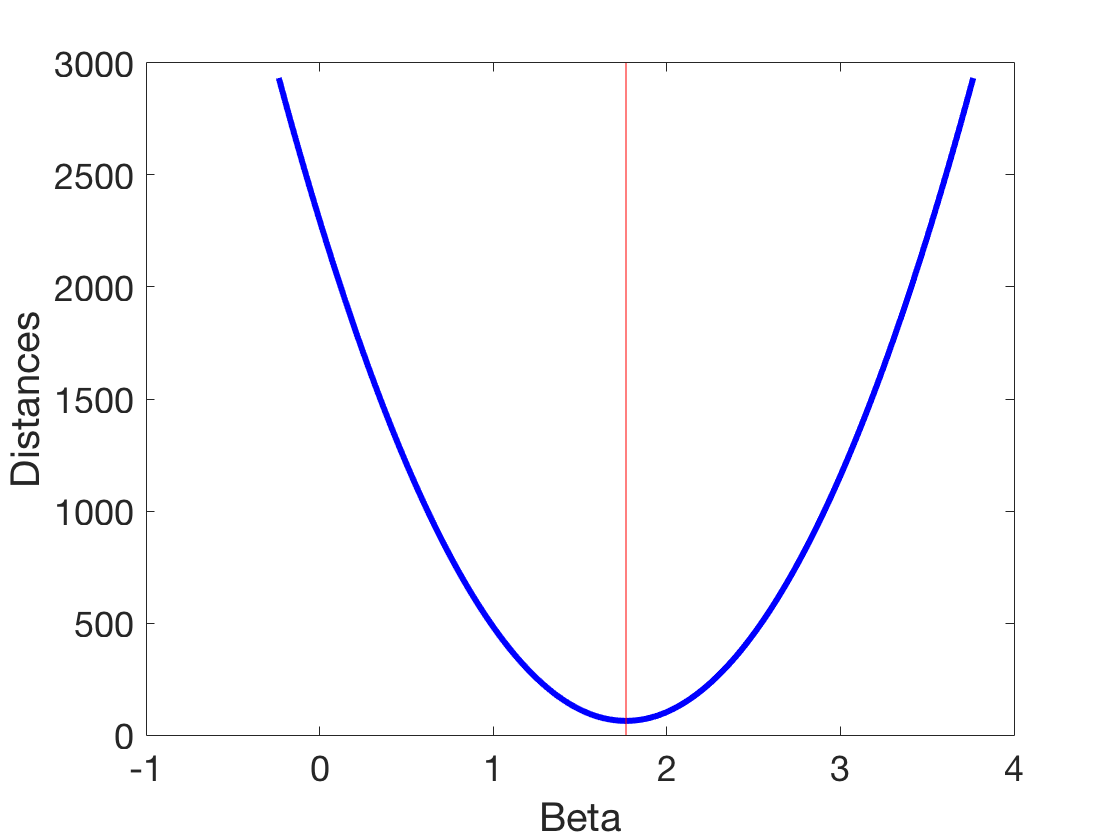
Going deeper into optimization - and gradient descent
beta startExploration = beta-2; endExploration = beta+2; testBetas = linspace(startExploration,endExploration,200); for ii = 1:length(testBetas) prediction(:,ii) = testBetas(ii)*xLuminance; distanceSum(1,ii) = sum(prediction(:,ii)-ySpikes); %Simple distanceSum(2,ii) = sum((prediction(:,ii)-ySpikes).^2); %Squared distanceSum(3,ii) = sum(abs(prediction(:,ii)-ySpikes)); %Absolute value distanceSum(4,ii) = sum(log(1 + (prediction(:,ii)-ySpikes).^2)); %Lorentzian distanceSum(5,ii) = sum(log(prediction(:,ii)-ySpikes)); %Weirdness distanceSum(6,ii) = sum(log(prediction(:,ii)-ySpikes).^2); %Weirdness end figure plot(testBetas,distanceSum(2,:),'b','linewidth',3) line([beta beta],[min(ylim) max(ylim)],'color', 'r') xlabel('Beta') ylabel('Distances') set(gca,'fontsize',18) figure for ii = 1:6 subplot(2,3,ii) plot(testBetas,distanceSum(ii,:),'b','linewidth',3) line([beta beta],[min(ylim) max(ylim)],'color', 'r') xlabel('Beta') ylabel('Distances') if ii == 1 title('Summed differences') elseif ii == 2 title('Sum of squared differences') elseif ii == 3 title('Sum of absolute differences') elseif ii == 4 title('Lorentzian?') elseif ii == 5 title('Sum of log differences') elseif ii == 6 title('Sum of squared log differences') end set(gca,'fontsize',18) end set(gcf,'color','w')
beta =
1.76497701782015
Warning: Imaginary parts of complex X and/or Y arguments ignored
Warning: Imaginary parts of complex X and/or Y arguments ignored
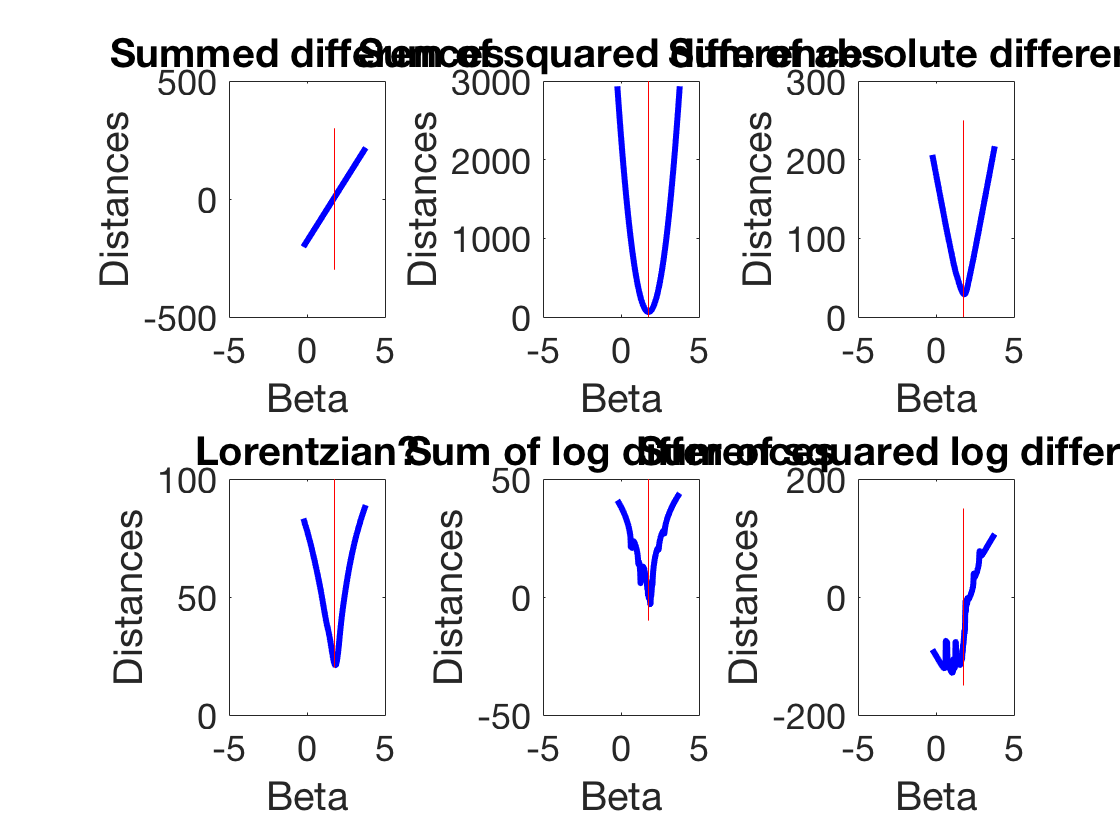
Gradient descent revisited
figure for ii = 1:length(testBetas) subplot(1,2,1) hold on prediction = testBetas(ii)*xLuminance; h7 = plot(xLuminance, ySpikes, 'o'); set(h7,'markersize',12) set(h7,'markerfacecolor','b') set(h7,'markeredgecolor','k') xlabel('Luminance in cd/m^2') ylabel('Spikes in ips') set(gca,'fontsize',18) regressionLineX = linspace(0,10,100); %Intrapolate regressionLineY = testBetas(ii) * regressionLineX; h8 = plot(regressionLineX, regressionLineY, 'k'); set(h8,'linewidth',2); for jj = 1:length(ySpikes) h(jj) = line([xLuminance(jj) xLuminance(jj)],[prediction(jj) ySpikes(jj)],'color','r'); end ylim([-5 1.5*max(ySpikes)]) subplot(1,2,2) hold on plot(testBetas(ii),distanceSum(2,ii),'.','color','b','linewidth',3) xlabel('Beta') ylabel('Distance') set(gca,'fontsize',18) xlim([startExploration endExploration]) ylim([0 max(distanceSum(2,:))]) shg pause(0.1) if ii < length(testBetas) delete(h8) delete(h) end end
PCA
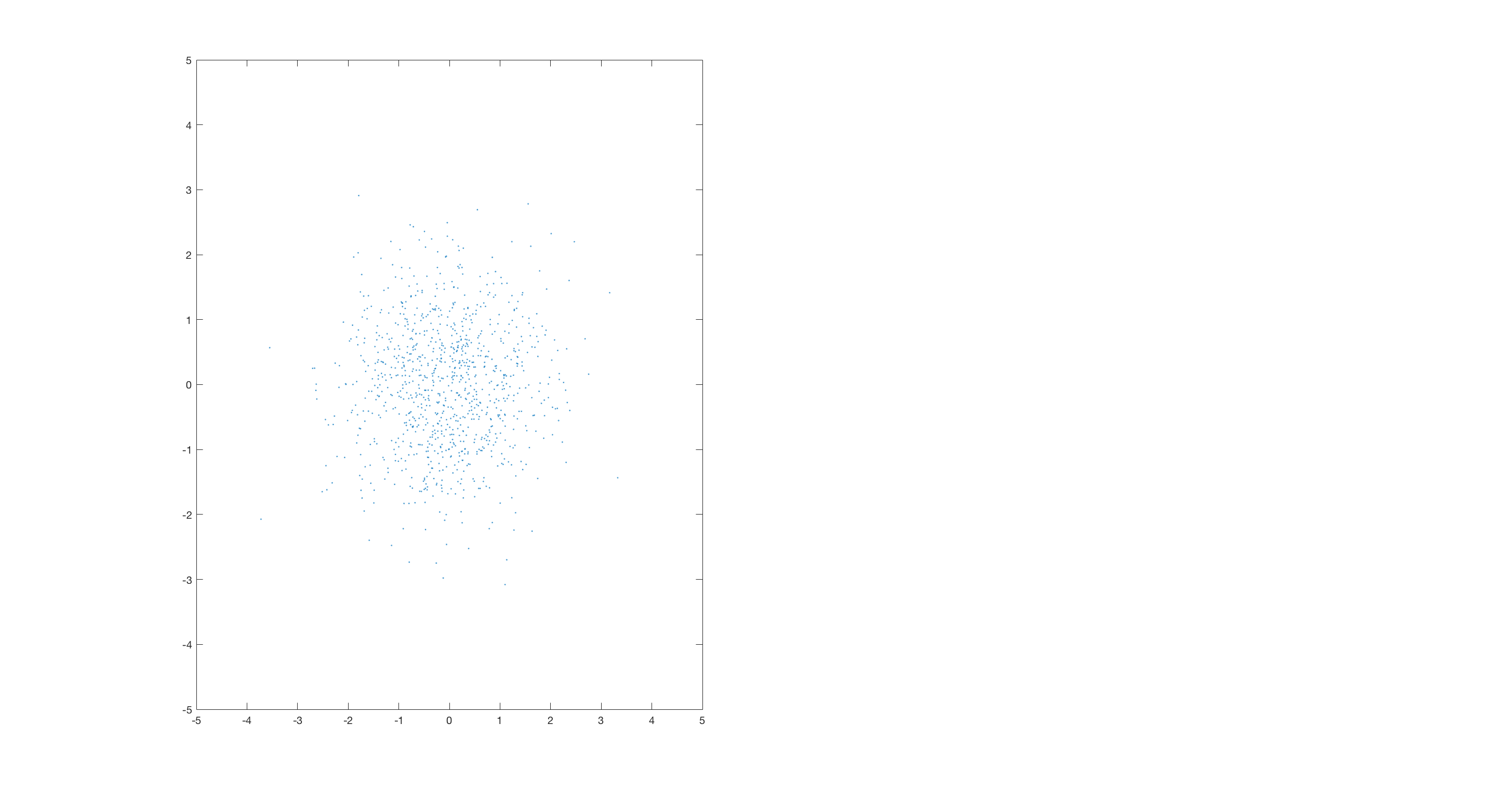
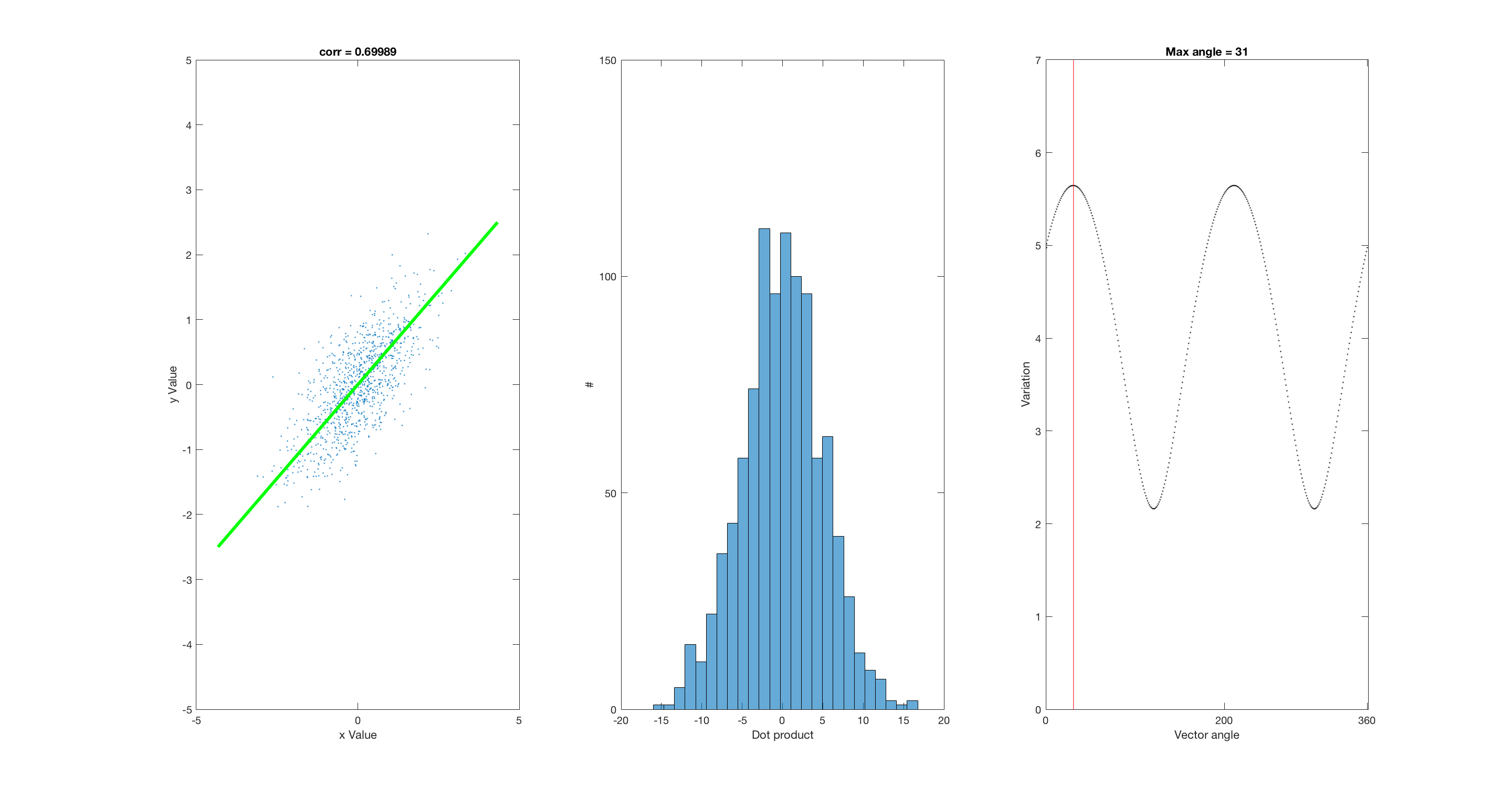
corr = 0.7 %Possible values: 0, 0.7, 0.9, 0.3, -0.7 A = randn(1000,2); %uncorrelated data if corr == 0.7 % Corr ~ 0.7 B = randn(1000,1); C = randn(1000,1); D = (B + C)./2; %correlated data (B and D) elseif corr == 0 B = randn(1000,1); D = randn(1000,1); elseif corr == 0.9 B = randn(1000,1); C = randn(1000,1); D = (3.*B + C)./4; %correlated data (B and D) elseif corr == 0.3 Q = randn(1000,1); B = randn(1000,1); C = randn(1000,1); D = ((3*Q) + B + C)./5; %correlated data (B and D) elseif corr == -0.7 B = randn(1000,1); C = randn(1000,1); D = (B + C)./2; %correlated data (B and D) D = D.*-1; end dotVec = [B,D]; dotProducts = zeros(1000,1); spreads = zeros(360,1); fullScreen subplot(1,2,1) plot(A(:,1),A(:,2),'.') xlim([-5 5]) ylim([-5 5]) fullScreen subplot(1,3,1) plot(B(:,1),D(:,1),'.') xlabel('x Value') ylabel('y Value') temp = corrcoef(B,D); title(['corr = ', num2str(temp(1,2))]) xlim([-5 5]) ylim([-5 5]) M = [5;0]; hold on plotv(M,':'); plotv(-1.*M,':'); h = findobj('linestyle',':'); set(h,'color','r') set(h,'linestyle','-') for ii = 1:1000 dotProducts(ii,1) = dot(dotVec(ii,:),M); end subplot(1,3,2) histogram(dotProducts,25) xlabel('Dot product') ylabel('#') xlim([-20 20]) ylim([0 150]) subplot(1,3,3) plot(1,std(dotProducts),'.','color','k'); xlabel('Vector angle') ylabel('Variation') xlim([0 361]) ylim([0 7]) set(gca,'XTick', [0 200 360]) hold on %set(h,'color','r') %columnVector = [-5 5]' %hold on pause %R = [0 -1; 1 0]; for aa = 1:360 subplot(1,3,1) angle = deg2rad(aa); R = [cos(angle) -sin(angle); sin(angle), cos(angle)]; RM = R*M; set(h,'linestyle','none') plotv(RM,':'); plotv(-1.*RM,':'); h = findobj('linestyle',':'); set(h,'color','r') set(h,'linestyle','-') pause(0.005) for ii = 1:1000 dotProducts(ii,1) = dot(dotVec(ii,:),RM); end subplot(1,3,2) histogram(dotProducts,25) xlabel('Dot product') ylabel('#') xlim([-20 20]) ylim([0 150]) subplot(1,3,3) spreads(aa) = std(dotProducts); plot(aa,spreads(aa),'.', 'color','k'); xlabel('Vector angle') ylabel('Variation') set(gca,'XTick', [0 200 360]) hold on %columnVector2 = R*columnVector; %plot(columnVector2,'color','r') %myVector = [-5 0; 5 0]l %myVector2 = R*myVector; %line(myVector(:,1),myVector(:,2),'color','r') %pause %line(myVector2(:,1),myVector2(:,2),'color','b') end maxSpreads = max(spreads); temp = find(spreads == maxSpreads); title(['Max angle = ', num2str(temp(1))]) line([temp(1) temp(1)],ylim,'color','r') subplot(1,3,1) M = [5;0]; angle = deg2rad(temp(1,1)-1); R = [cos(angle) -sin(angle); sin(angle), cos(angle)]; RM = R*M; set(h,'linestyle','none') plotv(RM,':'); plotv(-1.*RM,':'); h = findobj('linestyle',':'); set(h,'color','g') set(h,'linestyle','-') set(h,'linewidth',3)
corr =
0.7